Plant immunity: plants are not defenceless
Innate immunity: a common concept in animals and plants
Conserved microbial signatures, so-called microbe-associated molecular patterns (MAMPs), betray the pathogen’s presence to the host. MAMP detection by specific host pattern-recognition receptors (PRRs) is an integral part of the immune system of animals and plants. Cell surface components such as bacterial lipopolysaccharide (LPS), peptidoglycan and flagellin are typical MAMPs as they are vital for microbial survival and common to whole microbial classes. In mammals, MAMPs are sensed by different classes of PRRs e.g. the Toll-like receptors (TLRs), and trigger inflammatory responses. Vertebrates additionally evolved an adaptive immune system employing highly specific antibodies. Plants do not possess specialized roaming immune cells like vertebrates and rely solely on genetically determined innate immune responses that can be executed by almost all plant cells. In plants, sensing of MAMPs by PRRs induces a common set of signalling and defence responses known as pattern-triggered immunity (PTI). Despite some conceptual similarities, MAMP sensing apparently evolved independently in animals and plants.
Functions of lipopolysaccharide in host-bacteria interactions
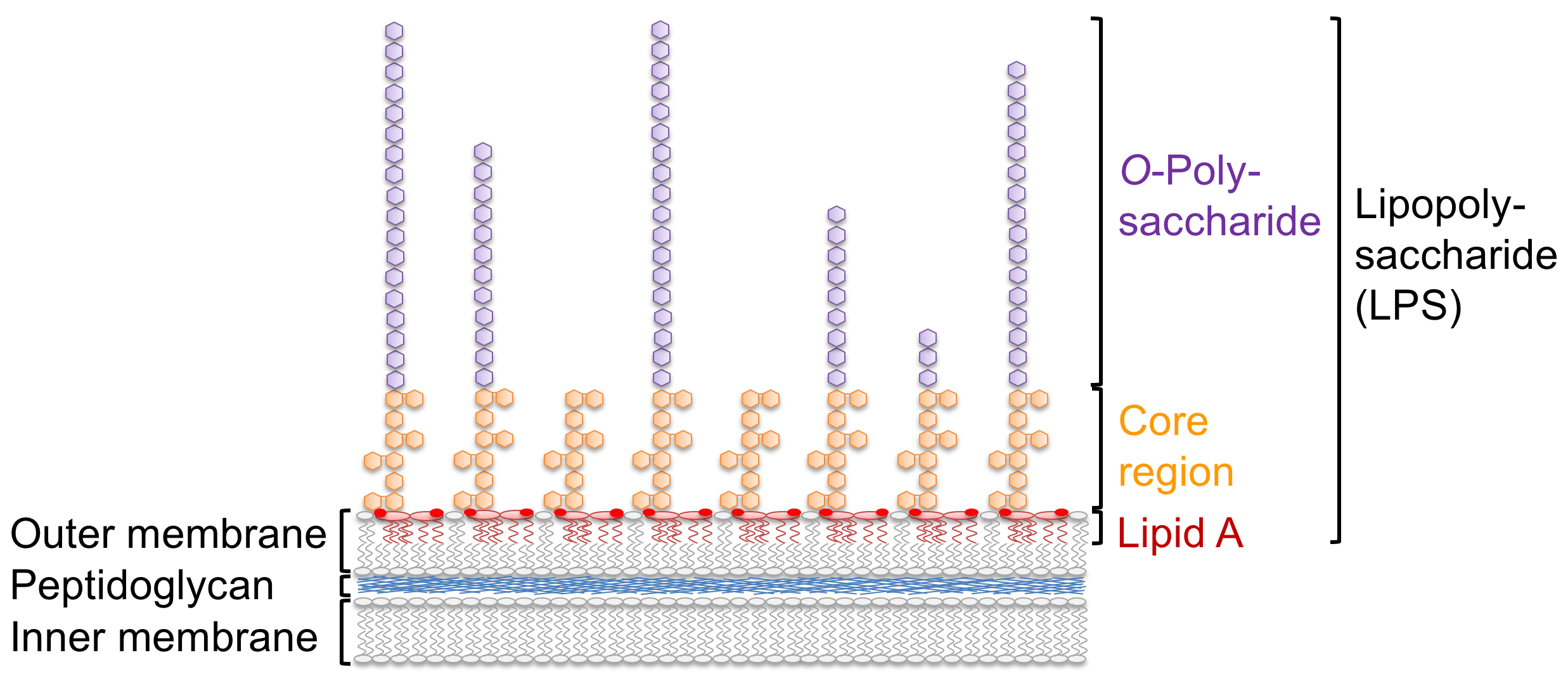
Lipopolysaccharide immune sensing differs in animals and plants
In mammals, LPS is a strong agonist of the innate immune system and its lipid A domain is sensed in trace amounts by extra- and intracellular immune receptors. LPS is also sensed in different plant species, but the perception systems are not yet understood. In a genetic screen for LPS sensing components in the model plant Arabidopsis thaliana, we identified the lectin S-domain receptor kinase LORE (LipoOligosaccharide-specific Reduced Elicitation) (Ranf et al., Nature Immunology, 2015). Interestingly, LORE does not sense LPS itself but free 3-hydroxylated fatty acids of medium chain length, which apparently co-purify with LPS during extraction from bacterial cell material. In Pseudomonas, medium chain 3-hydroxy fatty acids are part of the lipid A moiety of LPS and released during synthesis of penta-acylated LPS in the outer membrane by the lipid A O-deacylase PagL. 3-hydroxy fatty acids are also building blocks of other bacterial compounds and are presumably produced through different metabolic pathways (Kutschera et al., Science, 2019). Hence, in contrast to mammals which sense complex structures such as the lipid A domain of LPS, Arabidopsis plants sense small, low-complexity 3-hydroxy fatty acid metabolites.
In humans (left panel), LPS is sensed by different immune cells through different extra- and intracellular receptors. LPS is disaggregated from the bacterial membrane by the serum protein LBP and transferred to CD14, which occurs as soluble (sCD14) and membrane-linked (mCD14) version. Dependent on the cell type, CD14 can trigger LPS signalling itself, or further transfers LPS to the membrane-resident TLR4/MD-2 receptor complex. Lipid A binding to a preformed TLR4/MD-2 hetero-dimer leads to association with another TLR4/MD-2-dimer and initiates intracellular signalling. Depending on the cellular localization (at the plasma membrane or in endosomes upon CD14-dependent endocytosis) TLR4/MD-2/LPS complexes activate production of either interferons or cytokines through distinct signalling adapters (TIRAP/MyD88 or TRIF/TRAM). Intracellular LPS leads to oligomerization of caspase-4, activation of the non-canonical inflammasome and pyroptotic cell death.
In Arabidopsis plants (right panel), the bulb-type lectin S-domain-1 receptor kinase LORE was identified in a genetic screen for LPS sensing components. LORE does not sense LPS directly but free medium-chain 3-hydroyx fatty acids, which are released from the LPS lipid A moiety and presumably other metabolic pathways. In analogy to other SD-RLKs, LORE likely forms dimers and is activated through mutual phosphorylation by the cytosolic kinase domain.
(Figure adapted from Ranf et al. 2016, PLoS Pathogens)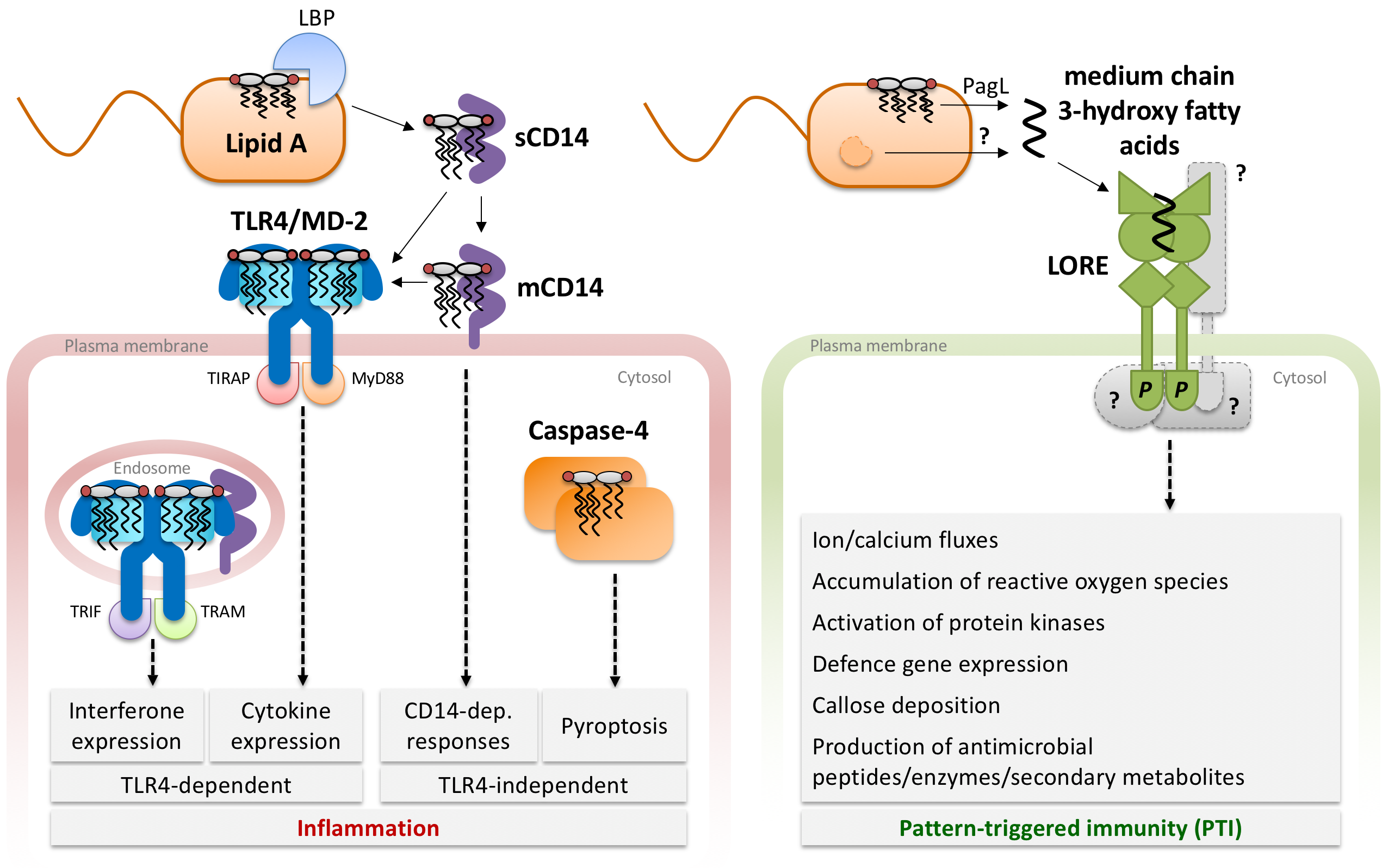
Funding

|

|

|
